-Search query
-Search result
Showing 1 - 50 of 154 items for (author: unge & j)

EMDB-19576: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-19582: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : LM32Cs3H1 sKO STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8rxh: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8rxx: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME WITH A/P/E-site tRNA AND mRNA : LM32Cs3H1 sKO STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17216: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8ovj: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17208: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17212: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

EMDB-17249: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A
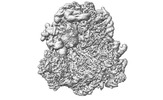
EMDB-17250: 
CRYO-EM FOCUSED REFINEMENT MAPS OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

EMDB-17254: 
CRYO-EM CONSENSUS MAP OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 SKO
Method: single particle / : RAJAN KS, YONATH A
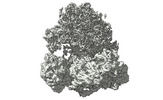
EMDB-17255: 
CRYO-EM FOCUSED REFINEMENT OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

PDB-8ova: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : PARENTAL STRAIN
Method: single particle / : Rajan KS, Yonath A

PDB-8ove: 
CRYO-EM STRUCTURE OF TRYPANOSOMA BRUCEI PROCYCLIC FORM 80S RIBOSOME : TB11CS6H1 snoRNA mutant
Method: single particle / : Rajan KS, Yonath A

EMDB-28615: 
MicroED structure of an Aeropyrum pernix protoglobin mutant
Method: electron crystallography / : Danelius E, Gonen T, Unge JT

PDB-8eum: 
MicroED structure of an Aeropyrum pernix protoglobin mutant
Method: electron crystallography / : Danelius E, Gonen T, Unge JT

EMDB-15272: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : snoRNA MUTANT
Method: single particle / : Rajan KS, Yonath A, Bashan A

PDB-8a98: 
CRYO-EM STRUCTURE OF LEISHMANIA MAJOR 80S RIBOSOME : snoRNA MUTANT
Method: single particle / : Rajan KS, Yonath A, Bashan A

EMDB-16457: 
Drosophila melanogaster Rab7 GEF complex Mon1-Ccz1-Bulli
Method: single particle / : Schaefer J, Herrmann E, Kuemmel D, Moeller A

EMDB-16462: 
MCB complex from drosophila melanogaster, local refinement map of the bottom part of the complex
Method: single particle / : Schaefer J, Herrmann E, Kuemmel D, Moeller A

EMDB-16463: 
MCB complex from drosophila melanogaster, local refinement map of the MCB core
Method: single particle / : Schaefer J, Herrmann E, Kuemmel D, Moeller A

EMDB-16464: 
MCB complex from drosophila melanogaster, consensus refinement
Method: single particle / : Schaefer J, Herrmann E, Kuemmel D, Moeller A

PDB-8c7g: 
Drosophila melanogaster Rab7 GEF complex Mon1-Ccz1-Bulli
Method: single particle / : Schaefer J, Herrmann E, Kuemmel D, Moeller A

EMDB-28616: 
MicroED structure of an Aeropyrum pernix protoglobin metallo-carbene complex
Method: electron crystallography / : Danelius E, Gonen T, Unge JT

PDB-8eun: 
MicroED structure of an Aeropyrum pernix protoglobin metallo-carbene complex
Method: electron crystallography / : Danelius E, Gonen T, Unge JT

EMDB-28947: 
Cryo-EM structure of the SARS-CoV-2 HR1HR2 fusion core complex with N969K mutation
Method: single particle / : Yang K, Brunger AT

EMDB-28948: 
Cryo-EM structure of the SARS-CoV-2 Omicron HR1-42G complex
Method: single particle / : Yang K, Brunger AT

PDB-8fa1: 
Cryo-EM structure of the SARS-CoV-2 HR1HR2 fusion core complex with N969K mutation
Method: single particle / : Yang K, Brunger AT

PDB-8fa2: 
Cryo-EM structure of the SARS-CoV-2 Omicron HR1-42G complex
Method: single particle / : Yang K, Brunger AT

EMDB-14964: 
HOPS tethering complex from yeast, composite map
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14965: 
HOPS tethering complex from yeast, consensus map covering the upper part of the complex
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14966: 
HOPS tethering complex from yeast, consensus map covering the bottom part of the complex
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14967: 
HOPS tethering complex from yeast, local refinement map of the SNARE-binding module
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14968: 
HOPS tethering complex from yeast, local refinement map of the backbone part of the complex
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14969: 
HOPS tethering complex from yeast, local refinement map of the bottom part of the complex (Vps18)
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

EMDB-14970: 
HOPS tethering complex from yeast, local refinement map of the bottom part of the complex (Vps39)
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C

PDB-7zu0: 
HOPS tethering complex from yeast
Method: single particle / : Shvarev D, Schoppe J, Koenig C, Perz A, Fuellbrunn N, Kiontke S, Langemeyer L, Januliene D, Schnelle K, Kuemmel D, Froehlich F, Moeller A, Ungermann C
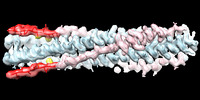
EMDB-27098: 
Cryo-EM structure of the SARS-CoV-2 HR1HR2 fusion core complex with extended HR2
Method: single particle / : Yang K, Brunger AT

PDB-8czi: 
Cryo-EM structure of the SARS-CoV-2 HR1HR2 fusion core complex with extended HR2
Method: single particle / : Yang K, Brunger AT

EMDB-25456: 
MicroED structure of proteinase K from a 130 nm thick lamella measured at 120 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25457: 
MicroED structure of proteinase K from a 200 nm thick lamella measured at 120 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25458: 
MicroED structure of proteinase K from a 325 nm thick lamella measured at 120 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25459: 
MicroED structure of proteinase K from a 115 nm thick lamella measured at 200 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25460: 
MicroED structure of proteinase K from a 130 nm thick lamella measured at 200 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25461: 
MicroED structure of proteinase K from a 95 nm thick lamella measured at 200 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25462: 
MicroED structure of proteinase K from a 540 nm thick lamella measured at 200 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25463: 
MicroED structure of proteinase K from a 460 nm thick lamella measured at 200 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25464: 
MicroED structure of proteinase K from a 260 nm thick lamella measured at 200 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25465: 
MicroED structure of proteinase K from a 530 nm thick lamella measured at 200 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB

EMDB-25466: 
MicroED structure of proteinase K from a 150 nm thick lamella measured at 300 kV
Method: electron crystallography / : Martynowycz MW, Clabbers MTB
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
